An Introductory Tutorial to the SEEKR2 (Simulation Enabled Estimation of Kinetic Rates v. 2) Multiscale Milestoning Software [Article v1.0]
DOI:
https://doi.org/10.33011/livecoms.5.1.2359Keywords:
SEEKR2, drug-target kinetics, residence time, Brownian dynamics, molecular dynamics, Markovian milestoning, multiscale simulationAbstract
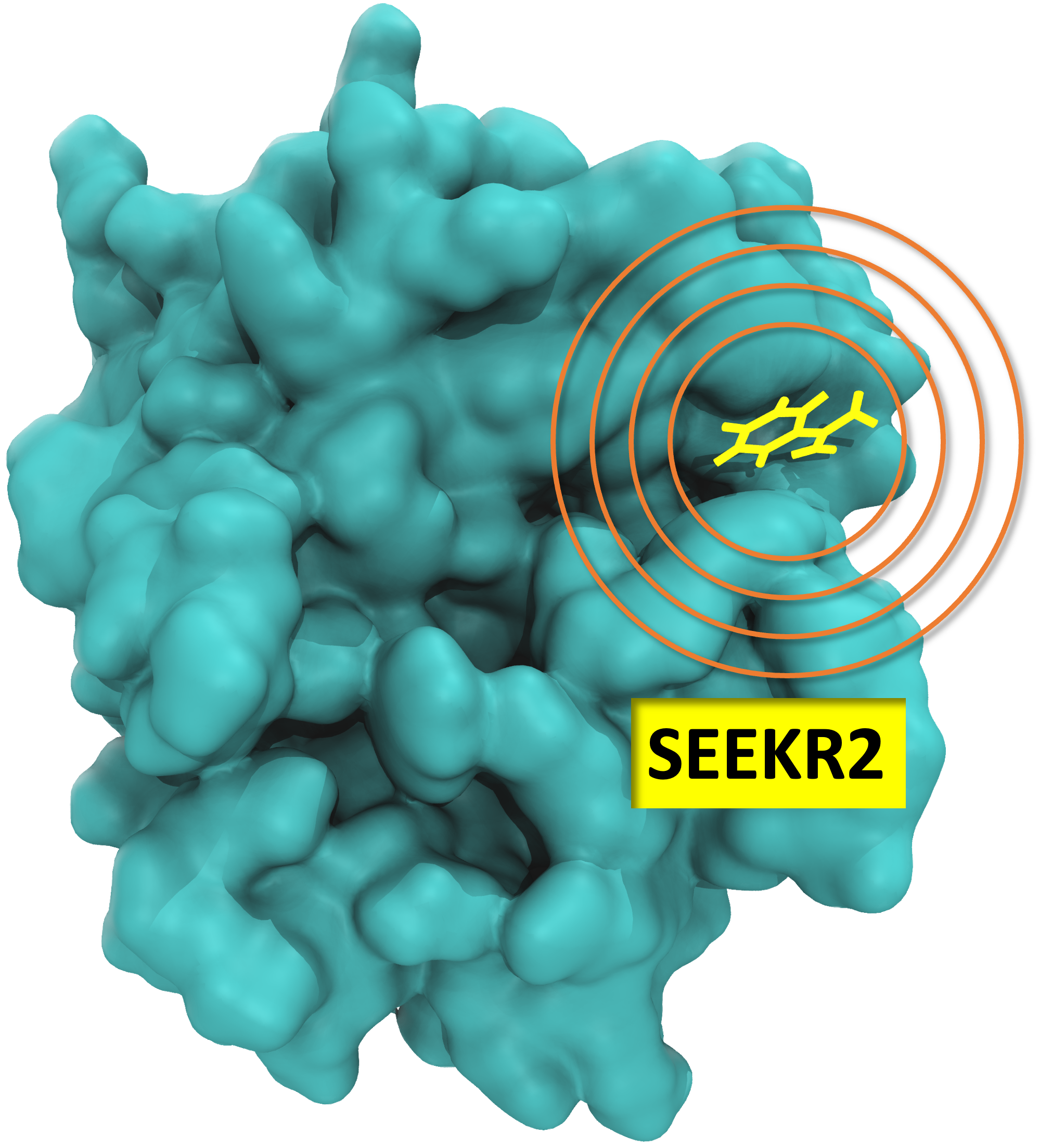
SEEKR2 (Simulation enabled estimation of kinetic rates v. 2) is a powerful and versatile software tool designed to computationally estimate the kinetics and thermodynamics of complex molecular processes, particularly emphasizing the process of receptor-ligand binding and unbinding. We present a suite of tutorials for the SEEKR2 (Simulation enabled estimation of kinetic rates v. 2) multiscale milestoning software. This tutorial presents a comprehensive guide for users offering the best practices for preparing, executing, and analyzing molecular dynamics (MD) and Brownian dynamics (BD) simulations using SEEKR2. This tutorial highlights the advancements presented in SEEKR2 - the latest iteration within the SEEKR programs, including significant improvements in speed and capabilities compared to its earlier versions. SEEKR2 now supports both NAMD and OpenMM simulation engines, providing users with more flexibility in their simulation setups. Additionally, the BD component has been upgraded to the Browndye2 engine, enhancing the accuracy and efficiency of simulations. This tutorial aims to guide users to install SEEKR2, run MD and BD simulations within the framework of the SEEKR2 program, and analyze and interpret the kinetics and thermodynamics of binding and unbinding of model host-guest systems, thereby demonstrating its ease of usability and extensible features that allow for future expansions of the method. This tutorial equips users with the necessary knowledge to effectively prepare, execute, and analyze simulations using SEEKR2. By following the best practices outlined in the tutorial, users can leverage the power of the SEEKR2 program to gain insights into complex molecular processes and accelerate their understanding of key biomolecular interactions.
Downloads
Published
How to Cite
License
Copyright (c) 2024 Anupam Anand Ojha, Lane William Votapka, Gary Alexander Huber, Shang Gao, Rommie Elizabeth Amaro

This work is licensed under a Creative Commons Attribution 4.0 International License.

