Kinase Similarity Assessment Pipeline for Off-Target Prediction [Article v1.0]
DOI:
https://doi.org/10.33011/livecoms.3.1.1599Abstract
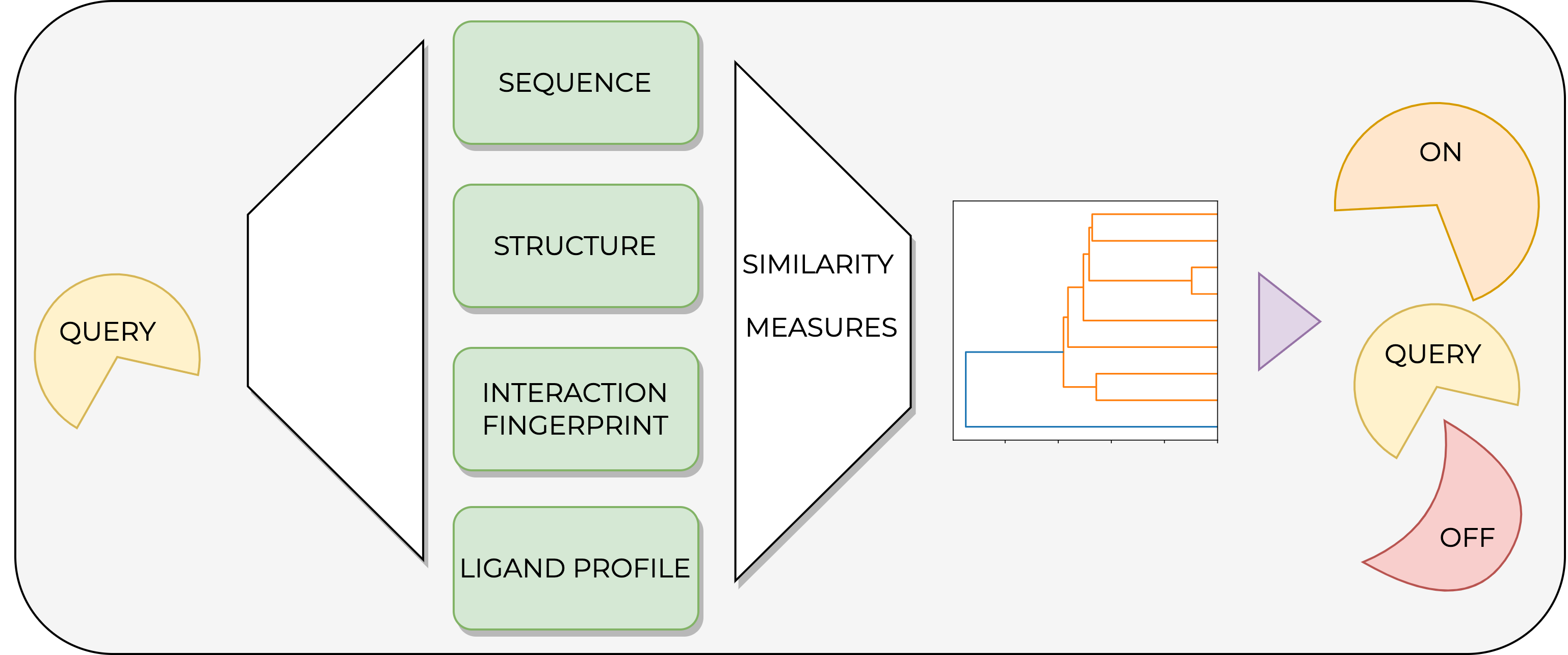
Kinases are established drug targets to combat cancer and inflammatory diseases. Despite decades of kinase research, challenges still remain, such as the under-exploration of a large fraction of the kinome and the promiscuous binding of many kinase inhibitors. Due to the highly conserved orthosteric ATP binding site in kinases, ligands may bind not only to their designated kinase (on-target) but also to other kinases (off-targets). Such promiscuous binding can cause mild to severe side effects, and the prediction of these off-targets is highly non-trivial. Therefore, we propose a pipeline that allows the study of kinase similarities from four different angles in an automated and modular fashion. The first method considers the binding site sequence. The second method uses structural information via KiSSim, a newly developed fingerprint that considers both physico-chemical and spatial properties of the binding site. The third method involves kinase-ligand interaction fingerprints as provided by KLIFS, and the last method utilizes the measured activity of ligands on kinases based on ChEMBL data. Finally, results for a given set of kinases are collected and analyzed to gain insight into potential off-targets from the different aforementioned perspectives. Since the pipeline is set up as a series of Jupyter notebooks covering both theoretical and practical aspects, the target audience ranges from beginners to advanced users working in the field of natural and computer sciences. The pipeline is part of the TeachOpenCADD project and extends it with this special kinase edition. All code is free, open-source, and made available at https://github.com/volkamerlab/teachopencadd.
Downloads
Published
How to Cite
License
Copyright (c) 2022 Talia Kimber, Dominique Sydow, Andrea Volkamer

This work is licensed under a Creative Commons Attribution 4.0 International License.

