Lessons learned in atomistic simulation of double-stranded DNA: Solvation and salt concerns [Article v1.0]
DOI:
https://doi.org/10.33011/livecoms.1.2.9974Keywords:
amber, molecular dynamics, nucleic acids, sampling, force fieldAbstract
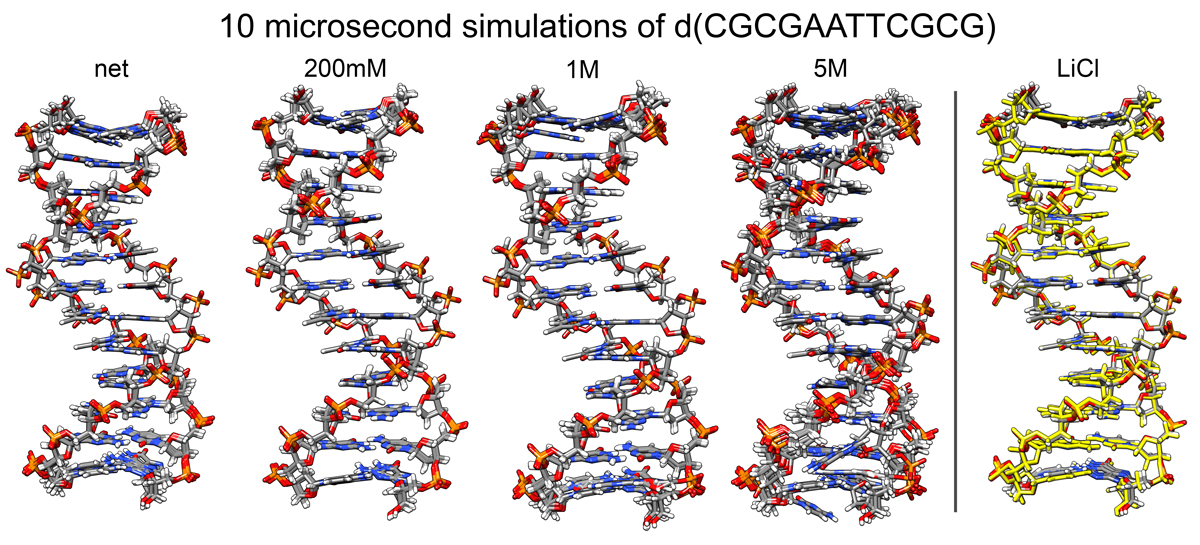
Nucleic acids are highly charged macromolecules sensitive to their surrounding of water, salt, and other charged molecules. Molecular dynamics simulations provide an atomistic view on DNA and RNA that has been useful to study the structure and dynamics of these molecules and their biological relevance. In this work we study the Drew-Dickerson dodecamer with the sequence GCGCAATTGCGC in three different salt concentrations and using different salt types to detect possible structural in2uence. Overall, the DNA shows no major structural change regardless of amount or type of ions used. Our results show that only at very high salt conditions (5M) a small structural effect is observed in the DNA duplex, which mainly consist of narrowing of the minor groove due to increased residence of ions. We also present the importance of sampling time to achieve a converged ensemble, which is of major relevance in any simulation to avoid biased or non-meaningful results.